- Research
- Open access
- Published:
Dissemination of clinical Escherichia coli strains harboring mcr-1, blaNDM−7 and siderophore-producing plasmids in a Chinese hospital
Antimicrobial Resistance & Infection Control volume 13, Article number: 66 (2024)
Abstract
Background
Carbapenem-resistant E. coli (CREco) pose a significant public health threat due to their multidrug resistance. Colistin is often a last-resort treatment against CREco; however, the emergence of colistin resistance gene mcr-1 complicates treatment options.
Methods
Two E. coli strains (ECO20 and ECO21), recovered from hospitalized patients in distinct wards, exhibited resistance to carbapenems and colistin. Whole-genome sequencing and phenotypic characterization were employed to study resistance patterns, plasmid profiles, transferability of resistance and virulence genes, and siderophore production capabilities. Comparative genome analysis was used to investigate the genetic environment of mcr-1, blaNDM−7, and virulence clusters.
Results
Both E. coli strains exhibited thr presence of both mcr-1 and blaNDM−7 genes, showing high resistance to multiple antibiotics. Genomic analysis revealed the clonal transmission of these strains, possessing identical plasmid profiles (pMCR, pNDM, and pVir) associated with colistin resistance, carbapenem resistance, and virulence factors. Conjugation experiments confirmed the transferability of these plasmids, indicating their potential to disseminate resistance and virulence traits to other strains. Comparative genomic analyses unveiled the distribution of mcr-1 (IncX4-type) and blaNDM (IncX3-type) plasmids across diverse bacterial species, emphasizing their adaptability and threat. The novelty of pVir indicates its potential role in driving the evolution of highly adaptable and pathogenic strains.
Conclusions
Our findings underscore the co-occurrence of mcr-1, blaNDM−7, and siderophore-producing plasmids in E. coli, which poses a significant concern for global health. This research is crucial to unravel the complex mechanisms governing plasmid transfer and recombination and to devise robust strategies to control their spread in healthcare settings.
Introduction
Carbapenem-resistant Enterobacterales (CRE), known for their multidrug resistance or extensively drug-resistant profiles, pose a severe threat to public health due to their association with high mortality and high transmissibility [1, 2]. With limited treatment options for CRE infections, colistin—an essential cationic antimicrobial peptide with neurotoxic effects—is considered the final resort against CRE [3]. However, the emergence and global dissemination of the plasmid-borne colistin resistance gene, mcr-1, presents a looming clinical crisis, potentially leading to a scarcity of effective antibiotics [4].
Carbapenemases serve as the primary cause of carbapenem resistance in CRE. KPC enzymes commonly prevail in Klebsiella pneumoniae, whereas NDM enzymes predominate in Escherichia coli [5]. Beyond their detection in hospitalized patients and healthy individuals, these enzymes are increasingly prevalent in diverse environments, including poultry, pets, water sources, and clinical settings [6, 7]. The rapid discovery of over 60 NDM variants since the identification of NDM-1 in India (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#NDM), suggests their adaptability, possibly fostering escalation among humans and food-producing animals. Similarly, since the first transferable mcr-1 was found in E. coli from China (2016), various mcr variants (mcr-1 to mcr-10) have been reported globally in diverse multidrug-resistant Enterobacterales, with mcr-1 emerging as the most prevalent variant [8]. Iron, crucial for bacterial growth and reproduction, prompts certain bacteria to secrete high-affinity siderophores like aerobactin and salmochelin, aiding their growth and pathogenicity [9, 10]. Notably, aerobactin genes iucABCD-iutA, residing on virulence plasmids, are considered pivotal pathogenic factors in K. pneumoniae [9], although reports of such plasmids in E. coli are relatively infrequent.
Plasmids, serving as carriers for genes like mcr-1, blaNDM, and the virulence factor iucABCD-iutA, pose a significant threat by facilitating their transmission among humans, animals, and the environment. This dissemination could culminate in larger, potentially uncontrollable global outbreaks, amplifying clinical treatment pressures. Therefore, the imperative is the control of plasmid spread at its source to manage carbapenem-resistant E. coli, colistin-resistant E. coli, and siderophore-producing E. coli.
E. coli concurrently carrying mcr-1, blaNDM, and iucABCD-iutA genes possess the potential to confer carbapenem, polymyxin resistance, and siderophore production, paving the way for the emergence of more menacing pathogens. Our study, isolating two strains of E. coli from hospital environments bearing mcr-positive, blaNDM-positive, and virulence plasmids, underscores their self-transmissible nature, capable of transferring both resistance and pathogenicity individually or simultaneously to other strains. Immediate measures to control the spread of such strains are imperative.
Materials and methods
Bacterial strains
Two clinical Escherichia coli strains (ECO20 and ECO21) which were resistant to carbapenems and colistin, were identified from previous daily work and were stored at − 80 °C in 10% glycerol. After subculture on blood agar plates for 24 h, isolates were identified by MALDITOF/MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry). Carbapenem resistance genes and colistin genes were detected by amplification and sequencing. Detailed anonymized metadata, such as patients’ gender, age, dates of sample collection, sample types, and hospitalization days, were recorded for analysis.
Antimicrobial susceptibility testing
We conducted antimicrobial susceptibility testing by the broth microdilution method using 0.5 McFarland inoculum suspensions diluted 1:100 to a final concentration of 106 CFU/ml. Ampicillin (AMP), ampicillin/sulbactam (SAM), piperacillin/tazobactam (TZP), cefazolin (CZO), cefuroxime (CXM), ceftriaxone (CRO), cefepime (FEP), cefoperazone/sulbactam (CSL), gentamicin (GEN), levofloxacin (LVX), trimethoprim/sulphamethoxazole (SXT), imipenem (IPM), meropenem (MEM), tigecycline (TGC), nitrofurantoin (NIT), amikacin (AMK), and colistin (COL) were selected to determine the minimum inhibitory concentrations (MICs). MICs were defined as the lowest concentrations of antimicrobials in the wells where no visible bacterial growth was observed. E. coli ATCC 25,922 served as a standard reference strain.
Compete genome sequencing and bioinformatics analysis
The genomic DNA of ECO20 and ECO21 isolates were extracted using the bacteria DNA isolation mini kit (Tiangen, Shanghai, China). Combined Oxford Nanopore (MinION system, Nanopore) and Illumina sequencing (NovaSeq system, Illumina Inc) were used to obtain complete chromosome and plasmid sequences. De novo assembly was performed using HGAP and CANU using default settings [11, 12]. The multilocus sequence type, serotype, and fimtype were determined using Center for Genomic Epidemiology (CGE) web tool (https://www.genomicepidemiology.org/services/). The virulence genes were identified using VFDB (http://www.mgc.ac.cn/cgi-bin/VFs/blast/blast.cgi), and antimicrobial resistance genes were collected using CARD (https://card.mcmaster.ca/analyze/rgi). Plasmid replicon type was determined by using Plasmid Finder 2.1 from the CGE. The conjugal transfer elements were identified using oriTfinder web-based tool (https://tool-mml.sjtu.edu.cn/oriTfinder/oriTfinder.html). Comparisons of pMCR, pNDM, and other similar plasmids were analysed using the BLAST Ring Image Generator (BRIG) software (http://sourceforge.net/projects/brig). Since the pMCR or pNDM mapped to many genomes, the top ten genomes were selected based on the BLAST scores to perform genome comparisons. Comparisons of iuc and iro clusters, and genetic environments of blaNDM−7 and mcr1.1 were performed using GBKviz (https://gbkviz.streamlit.app/).
Plasmid conjugation experiment
To investigate the transferabilities of blaNDM−7, mcr1.1, and virulence factors, conjugation experiments were performed between ECO20 and sodium azide-resistant E. coli J53. Both donor and recipient isolates were cultured to logarithmic and mixed in equal volumes and incubated at 37℃ overnight to facilitate conjugation. The agar containing sodium azide (80ug/ml) was used to calculate the number of recipients. Colistin (0.5 ug/ml) was used to screen transconjugants containing mcr1.1. Meropenem (0.25 ug/ml) was used to screen transconjugants containing blaNDM−7. Gentamicin (0.5 ug/ml) were used to screen transconjugants containing iuc clusters. The agar containing sodium azide and colistin or meropenem or gentamicin were used to screen transconjugants carrying blaNDM−7 or mcr-1or iuc. The presence of blaNDM−7, mcr, and iuc in transconjugants was confirmed by PCR and corresponding resistance phenotyping. The primers (irp3, blaNDM−7, mcr-1, and iutA) were provided in Table S1. Frequencies of conjugation transfer were calculated by the number of transconjugants per recipient. The experiments were repeated five times. S1 nuclease-Pulsed-Field Gel Electrophoresis (S1-PFGE) was further performed to confirm the plasmids in transconjugants. Genomic DNA digested with S1 nuclease was subjected to PFGE as described previously [13].
Chrome azurol S (CAS) agar plates
Chrome Azurol S (CAS) agar plates were utilized for qualitative detection of siderophore production. To prepare the CAS solution, 60.5 mg of CAS was dissolved in 50 mL of ddH2O. Subsequently, 1 mmol FeCl3·6H2O dissolved in 10 mL of 10 mM hydrochloric acid was added to this solution. After thorough mixing, the resultant mixture was slowly poured into a solution of acetyltrimethylammonium bromide (CTAB) (72.9 mg CTAB dissolved in 40 mL ddH2O), followed by high-pressure sterilization. The CAS-containing King’s B medium was prepared by autoclaving an appropriate amount of King’s B medium agar powder in 700 mL ddH2O. To this medium at a temperature of 50 °C ~ 60 °C, 200 mL of 10×PIPES buffer and 100 mL of the CAS solution were added. This medium was poured slowly onto plates and layered with LB agar that had been sterilized under high pressure to form CAS double-layer plates. The strains were cultured in a low-nutrient broth to a logarithmic phase and the concentration was adjusted to 105 CFU/ml. Subsequently, 1 µL bacterial culture was inoculated onto these plates. The plates were then incubated at 37 °C for 36 to 48 h, and observation was made for the formation of orange-yellow transparent rings.
Chrome azurol S (CAS) assay
A single fresh colony was inoculated into LB broth and incubated overnight at 37 °C. The following day, a 1:1000 dilution was made from this culture into low-nutrient broth and incubated overnight at 37 °C. Following centrifugation of 1 mL of the culture at 12,000 rpm for 10 min, the supernatant was collected. A CAS detection solution was prepared to achieve a final concentration of 1.2 mM CTAB, 2 mM CAS, and 1 mM FeCl3·6H2O. In triplicate, 100 µL of bacterial supernatant was added to a 96-well plate and mixed with an equal volume of the CAS detection solution. This mixture was incubated in darkness for 30 min, and the absorbance was measured at OD630 nm. Optical density values (As) were recorded using sterile low-nutrient broth as a reference (Ar). The siderophore activity unit Su (Siderophore unit) was calculated as [(Ar − As)/Ar]×100 [14]. The optical density values (As) were recorded, using a sterile low-nutrient broth as a reference (Ar). The experiment was repeated thrice. GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA) was used to assess the statistical significance of the data with the student’s t-test and the log-rank test.
Results
Identification and resistance phenotypes
Clinical E. coli strains (ECO20 and ECO21) were isolated from hospitalized patients at a hospital in Jining, Shandong. Notably, both strains exhibited the presence of both blaNDM−7 and mcr-1 genes. ECO20 was recovered from the urine sample of Patient 1 in the intensive care unit on February 17, 2021, while ECO21 was obtained from Patient 2’s sputum in the neurology department on February 19, 2021 (Figure S1). Despite their non-concurrent presence in the same ward, clinical records reveal a compelling temporal and spatial correlation. Patient 2, carrying ECO21, had a prior admission to the ICU ward from February 09 to February 18, 2021 (Figure S1). During an overlapping interval (February 15–18), both Patient 1 and Patient 2 coexisted within the ward, suggesting relevant temporal and spatial proximity influencing transmission dynamics.
Antimicrobial susceptibility testing revealed that both E. coli isolates conferred resistance to almost antimicrobials tested, including ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, cefazolin, cefuroxime, ceftriaxone, cefepime, cefoperazone/sulbactam, gentamicin, trimethoprim/sulphamethoxazole, imipenem, meropenem, and colistin, but no resistance to tigecycline, nitrofurantion, and amikacin (Table 1). Thus, these strains exhibited multidrug-resistant profiles.
Genomic characteristics
To further investigate the clonal relationship and genomic features, we sequenced and assembled their genomes. Notably, the ECO20 genome shared 99.99% Average Nucleotide Identity (ANI) with ECO21 (Figure S2), indicating the clonal transmission of these two isolates. The two strains belonged to ST4456, fimH20 and H4:O83, and harboring some common virulence genes such as csgA, fdeC, hlyF, ibeA, iss. They exhibited identical plasmid profiles: a 33 Kb pMCR plasmid, a 46 Kb pNDM plasmid, and a 175 Kb pVir plasmid (Table 2). The pMCR plasmid belonging to the IncX4 type carried the colistin resistance gene mcr-1, while the pNDM plasmid belonging to the IncX3 type carried the carbapenem resistance gene blaNDM−7. The pVir plasmid (IncFIB/IncFII type) harbored aerobactin and salmochelin-associated virulence genes, alongside additional resistance genes, including AAC(3)-IId, dfrA17, TEM-1, sul2, APH(3’’)-Ib, APH(6)-Id, and tet(A). Notably, all plasmids possessed conjugal transfer elements, albeit the oriT sequences of pNDM and pMCR were not identified by the oriTfinder web tool.
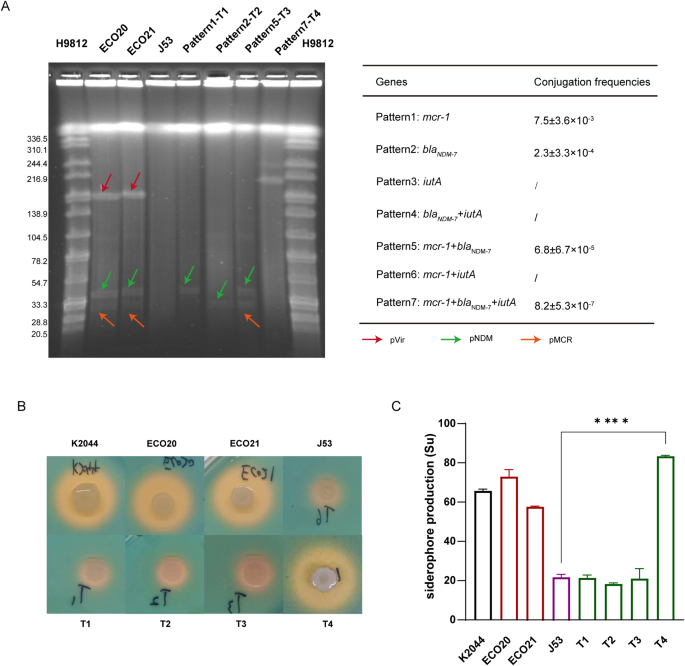
Transferability of resistance and virulence plasmids
To investigate the transmissibility of mcr-1, blaNDM−7, and iutA, we subjected ECO20 to conjugation experiments with E. coli J53. As shown in Fig. 1A, there should be seven plasmid conjugation patterns. However, after many conjugation experiments, we only observed four different transfer patterns (Fig. 1A). Importantly, the co-transfer of mcr-1, blaNDM−7, and iutA led to the formation of a fusion plasmid (Fig. 1A), suggesting that diverse recombination events would occur during the plasmid conjugation process. High conjugation frequencies of mcr-1 and blaNDM−7 indicate broad spread and effective transfer of pMCR and pNDM plasmids (Fig. 1A). The transfer of resistance genes mcr-1 and blaNDM−7 conferred their corresponding resistance phenotypes against colistin and carbapenem to E. coli J53 (Table 1). Similarly, virulence genes with their corresponding virulence phenotypes characterized by high siderophore production were also successfully transferred to E. coli J53. The transconjugant T4 which acquired the pVir plasmid from ECO20 showed significantly higher siderophore production than J53 (Fig. 1B and C).
Prevalence of pMCR and pNDM plasmids
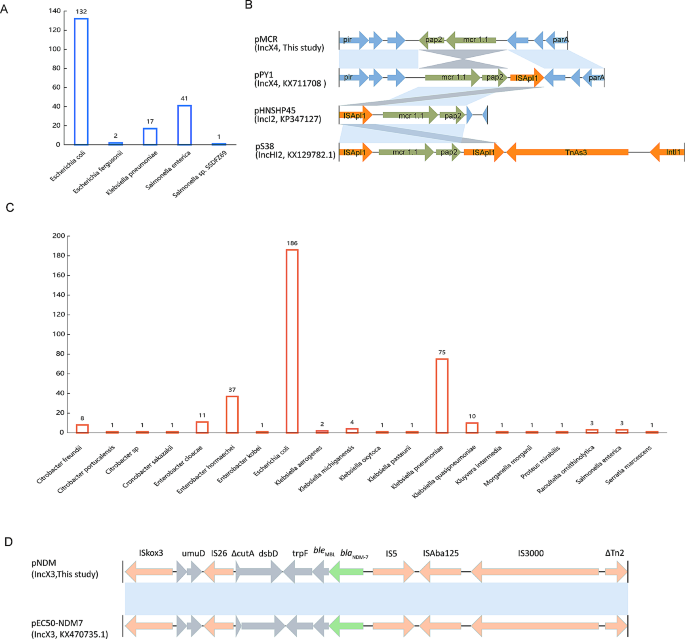
Blastn analysis revealed that the IncX4-type pMCR plasmid closely resembled numerous plasmids previously submitted to the NCBI database (Figure S3). These similar plasmids exhibited >99% coverages and >99% identities. Interestingly, it was not confined to E. coli but was also found in Klebsiella and Salmonella (Fig. 2A). Moreover, the single-ended Tn6330 variant (ISApl1-mcr1.1-pap2) is commonly present in mcr-1.1-harbouring plasmids, such as pHNSHP45(IncI2, KP347127) and pS38(IncHI2, KX129782.1), despite that ISApl1 was downstream of mcr1.1 and pap2 in pPY1(IncX4, KX711708) (Fig. 2B). In contrast, pMCR in this study had a different genetic environment for mcr1.1 with the pap2 located downstream and ISApl1 deleted (Fig. 2B).
The pNDM-like IncX3 plasmid was also identical to many plasmids from the NCBI database (Figure S4). These similar plasmids had 100 coverages and > 99% identities. They were frequently observed in gram-negative bacteria, exhibiting a broader host range compared to the pMCR-like IncX4. Apart from E. coli and Klebsiella, the pNDM-like IncX3 plasmid was also detected in Enterobacter, Citrobacter, and various other bacterial species (Fig. 2C). Nonetheless, E. coli was still the primary host bacterium carrying the blaNDM−7-positive IncX3 plasmid (Fig. 2C). Unlike the prevalence observed in pMCR-like IncX4, the incidence of pNDM-like IncX3 plasmids was lower in Salmonella (Fig. 2C). Furthermore, blaNDM−7 was associated with the complex transposon structure (IS26-IS3-ISAba125-IS5-blaNDM−7- bleMBL-trpF-dsbD-IS26) which is also commonly present in other blaNDM−7-positive plasmids and plasmids carrying NDM variants (Fig. 2D).
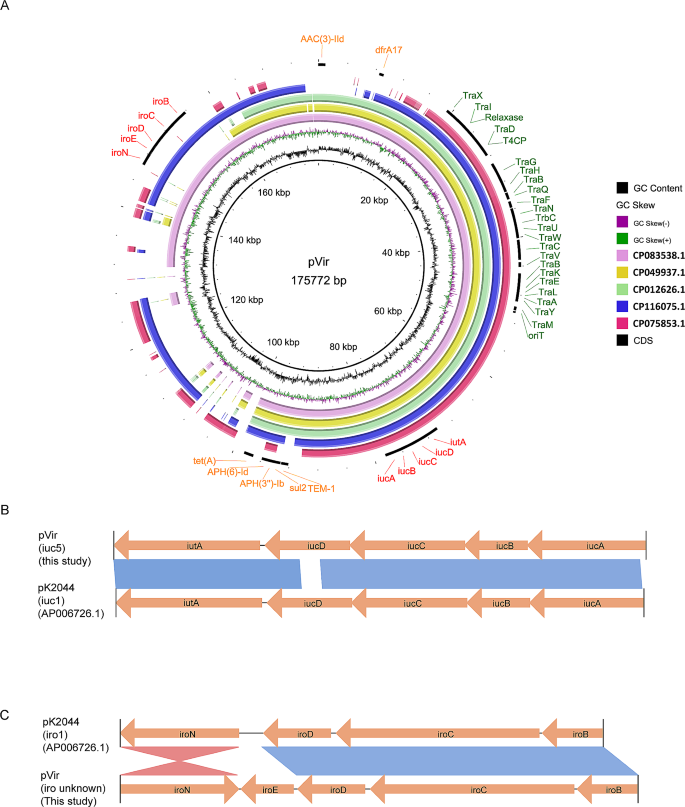
Characteristics of the IncFIBK/IncFII virulence plasmid
The 175,772 bp pVir plasmid contained virulence and resistance genes, along with a Type IV Secretion System (T4SS) (Fig. 3A). The aerobactin genes (iucABCD-iutA) and salmochelin genes (iroBCDEN) endowed it with high siderophore production (Fig. 1B and C). Notably, the schematic structure of iuc clusters (iuc1) was similar to that of virulence plasmid pK2044 in Klebsiella (iuc1) (Fig. 3B), while the iro clusters in pVir had iroE added and iroN inversion compared to pK2044 (Fig. 3C). The complete conjugal transfer element T4SS conferred the ability of self-transfer (Fig. 2A), and some resistance genes conferred resistance to aminoglycosides, cephalosporins, sulfonamides, tetracyclines, and diaminopyrimidines (Table 1). No identical plasmid was found in the NCBI nucleotide database. Some similar plasmids were selected to perform comparative genome analysis with pVir in this study. Most plasmids cannot possess all the above elements for self-transfer, resistance, and siderophore production, except for the unnamed plasmid (CP083538) (Fig. 3A).
Characteristics of siderophores-encoding pVir plasmids. (A) Comparative analysis of pVir plasmids and some similar plasmids from NCBI database. The reference plasmid is pVir (CP139892.1). (B) Comparisons of iuc and iro clusters carried by E. coli pVir plasmid and Klebsiella pneumoniae pK2044 plasmid
Discussion
The escalating threat of multidrug-resistant Enterobacterales (MDRE) represents a profound challenge in healthcare settings worldwide [15]. Our investigation highlights the emergence of carbapenemase genes (such as blaNDM) and the plasmid-mediated colistin resistance gene (mcr-1) within clinical E. coli strains. The coexistence of these resistance determinants, along with the concurrent presence of siderophore-producing plasmids, indicates a worrisome confluence that could lead to the emergence of multidrug-resistant and highly pathogenic pathogens.
The isolation of genetically identical strains (ECO20 and ECO21) carrying identical plasmid profiles from patients in distinct wards raises critical concerns about nosocomial transmission. The spatial and temporal correlation between patients suggests a possible route of transmission within the hospital environment. Notably, these strains exhibited extensive resistance profiles against a range of antibiotics, especially carbapenems and colistin, limiting treatment options in clinical settings. These findings resonate with the global trend in the dissemination of carbapenemases and colistin resistance in E. coli, as documented by recent studies [1, 4]. In contrast to NDM-1, NDM-7 has two amino acid substitutions (D130N and M15L) and is more active as a carbapenemase [16]. Its dissemination, mainly associated with the IncX3 conjugative plasmid [17], shares a genetic environment with most NDM variants, notably the IS26-flanked pseudo-composite transposons (IS26-IS3-ISAba125-IS5-blaNDM-bleMBL-trpF-dsbD-IS26) [18], which likely represents the most important contributor to the widespread of blaNDM genes. Our study finds that the IncX3-type pNDM plasmids, while observed across diverse gram-negative bacteria, exhibit preferential colonization in certain species, such as E. coli and Klebsiella. Their ability to traverse diverse ecological niches poses a significant public health threat.
The mcr gene exerts colistin resistance by encoding a phosphoethanolamine transferase and can be located on various plasmids, including the IncI2, IncX4, IncP, IncY, IncFII, and IncHI2 plasmid incompatibility clusters [19], which are considered to be important vectors for facilitating the spread of mcr-1. Among these, the most common plasmid types are IncI2, IncHI2, and IncX4 [19]. The mcr-1 in this study was located on conjugative IncX4 plasmids. While IncX4-type pMCR plasmids were found in a narrower range of bacterial species compared to IncX3-type pNDM plasmids, their prevalence was notably higher in Salmonella. This highlights distinct species-specific dissemination patterns observed for these resistance plasmids. The single-ended Tn6330 variant (ISApl1-mcr1.1-pap2) is commonly present in mcr1.1-harbouring plasmids [20], which drives the dissemination of mcr1.1 genes among various bacterial species. Even though plasmids can spread rapidly within species and across geographical regions, they aren’t very likely to establish themselves in new bacterial environments. Such exchanges, however, provide opportunities for between-plasmid transposon jumps and genetic recombination, which significantly contribute to the spread of antimicrobial resistance genes.
The co-occurrence of blaNDM-positive plasmid and mcr-positive plasmid possibly occurred, and several studies have been reported [21,22,23]. However, the co-existence of blaNDM−7 positive IncX3 plasmid and mcr1.1-positve IncX4 plasmid in clinical E. coli isolates which we described in this study was previously underreported. The IncX3 and IncX4 were the most common plasmids carrying blaNDM and mcr, respectively [17, 19], thus, plasmid transfer and evolution may allow the co-transfer of blaNDM and mcr to become prevalent and spread in the future.
Additionally, our study highlights the presence of a 175 kb pVir plasmid in ECO20 and ECO21, mediating high siderophore production via aerobactin and salmochelin encoding high-affinity siderophores (aerobactin and salmochelin) which are considered as critical virulence factors enabling bacteria to compete for iron with the host [9]. Adequate iron is crucial for bacteria to survive in limited environments [24]. The existing study focuses more on the siderophore-production K. pneumoniae for the rapid emergence of multidrug-resistant hypervirulent K. pneumoniae has attracted widespread attention [25, 26]. However, the importance of siderophore production extends beyond Klebsiella to encompass all pathogens. Notably, the iuc and iro clusters differed from those found in hypervirulent K. pneumoniae, suggesting a distinct ancestral background. The absence of identical plasmids in existing databases accentuates the novelty and significance of pVir, indicating its potential role in driving the evolution of highly adaptable and pathogenic strains.
Our results elucidate the conjugative abilities of blaNDM−7-positive IncX3 plasmid, mcr1.1-positive IncX4 plasmid, and iuc-positive IncFIB/IncFII plasmid, and describe four transfer patterns. Interestingly, the formation of fusion plasmid illustrates the potential genetic recombination during plasmid conjugation. Importantly, the transfer of resistance and virulence genes endows bacteria with corresponding resistance and virulence phenotypes, indicating its potential role in driving the evolution of highly adaptable and pathogenic strains. This emphasizes the urgency to prevent the spread of these plasmids and associated genes.
However, our study has limitations. The analysis was limited to two clinical isolates from a specific geographical location, restricting the generalizability of our findings. Moreover, the study has not detected all possible plasmid transfer patterns. Further studies involving a broader spectrum of isolates from diverse geographic regions are crucial to elucidate the extent of plasmid dissemination and genetic diversity.
Future research should delve into the intricate mechanisms underlying plasmid transfer and recombination, deciphering the molecular determinants governing these processes. Moreover, surveillance and control measures within healthcare settings are imperative to curb the spread of MDR E. coli strains and prevent the emergence of highly resistant and virulent strains. In conclusion, our study underscores the urgency of understanding the dynamics of plasmid-mediated resistance and virulence, emphasizing the need for robust surveillance and intervention strategies to mitigate the escalating threat posed by MDRE strains.
Data availability
The nucleotide sequences of the chromosome and plasmids of E. coli isolates ECO20 and ECO21 have been deposited in the NCBI database (BioProject accession number PRJNA1005813).
References
Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46.
Durante-Mangoni E, Andini R, Zampino R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin Microbiol Infect. 2019;25(8):943–50.
El-Sayed Ahmed MAE, Zhong LL, Shen C, Yang Y, Doi Y, Tian GB. Colistin and its role in the era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect. 2020;9(1):868–85.
Xiaomin S, Yiming L, Yuying Y, Zhangqi S, Yongning W, Shaolin W. Global impact of mcr-1-positive Enterobacteriaceae bacteria on one health. Crit Rev Microbiol. 2020;46(5):565–77.
Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314.
Ramírez-Castillo FY, Guerrero-Barrera AL, Avelar-González FJ. An overview of carbapenem-resistant organisms from food-producing animals, seafood, aquaculture, companion animals, and wildlife. Front Vet Sci. 2023;10:1158588.
Savin M, Bierbaum G, Mutters NT, Schmithausen RM, Kreyenschmidt J, García-Meniño I et al. Genetic characterization of Carbapenem-resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiot (Basel). 2022;11(4).
Hussein NH, Al-Kadmy IMS, Taha BM, Hussein JD. Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol Biol Rep. 2021;48(3):2897–907.
Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3).
Kramer J, Özkaya Ö, Kümmerli R. Bacterial siderophores in community and host interactions. Nat Rev Microbiol. 2020;18(3):152–63.
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–9.
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–36.
Tian D, Wang B, Zhang H, Pan F, Wang C, Shi Y et al. Dissemination of the bla (NDM-5) gene via IncX3-Type plasmid among Enterobacteriaceae in Children. mSphere. 2020;5(1).
Payne SM. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994;235:329–44.
Parmanik A, Das S, Kar B, Bose A, Dwivedi GR, Pandey MM. Current treatment strategies against Multidrug-resistant Bacteria: a review. Curr Microbiol. 2022;79(12):388.
Göttig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity. J Antimicrob Chemother. 2013;68(8):1737–40.
Shao C, Hao Y, Wang Y, Jiang M, Jin Y. Genotypic and phenotypic characterization of bla (NDM-7)-Harboring IncX3 plasmid in a ST11 Klebsiella pneumoniae isolated from a Pediatric patient in China. Front Microbiol. 2020;11:576823.
Weber RE, Pietsch M, Frühauf A, Pfeifer Y, Martin M, Luft D, et al. IS26-Mediated transfer of bla (NDM-1) as the Main Route of Resistance Transmission during a polyclonal, multispecies Outbreak in a German hospital. Front Microbiol. 2019;10:2817.
Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol. 2019;45(2):131–61.
Li R, Xie M, Lv J, Wai-Chi Chan E, Chen S. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother. 2017;72(3):696–9.
Wang X, Wang Y, Jiang X, Gong X, Wang Y, Shen Z. Co-transfer of mcr-8 with bla(NDM-1) or tmexCD1-toprJ1 by plasmid hybridisation. Int J Antimicrob Agents. 2022;60(2):106619.
Han H, Liu W, Cui X, Cheng X, Jiang X. Co-existence of mcr-1 and bla (NDM-5) in an Escherichia coli strain isolated from the Pharmaceutical Industry, WWTP. Infect Drug Resist. 2020;13:851–4.
Kuang X, Yang R, Ye X, Sun J, Liao X, Liu Y et al. NDM-5-Producing Escherichia coli Co-harboring mcr-1 gene in Companion animals in China. Anim (Basel). 2022;12(10).
Yilmaz B, Li H. Gut microbiota and Iron: the crucial actors in Health and Disease. Pharmaceuticals (Basel). 2018;11(4).
Yang X, Sun Q, Li J, Jiang Y, Li Y, Lin J, et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect. 2022;11(1):841–9.
Tian D, Liu X, Chen W, Zhou Y, Hu D, Wang W, et al. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect. 2022;11(1):1936–49.
Acknowledgements
Not applicable.
Funding
This work was supported by the China Postdoctoral Science Foundation (2022M721338), Shandong Postdoctoral Innovation Talents Support Program (SDBX2022052), Shandong Provincial Natural Science Foundation (ZR2023QH215), Jining Key Research and Development Program (2021YXNS025), Tai Shan Young Scholar Foundation of Shandong Province (tsqn202306400), Post-doctoral Program of Affiliated Hospital of Jining Medical University (JYFY321208), and Project of Shandong Province Higher Educational Youth Innovation Science and Technology Program (2022KJ106).
Author information
Authors and Affiliations
Contributions
DT and LL conceived and designed the work. MQ and LY collected and provided the isolates. MQ, AS, YT and XY performed the experiments and analyzed the data. DT drafted the manuscript. All the authors revised the manuscript and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, L., Zhao, M., Tang, Y. et al. Dissemination of clinical Escherichia coli strains harboring mcr-1, blaNDM−7 and siderophore-producing plasmids in a Chinese hospital. Antimicrob Resist Infect Control 13, 66 (2024). https://doi.org/10.1186/s13756-024-01423-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-024-01423-3